Proteins need to fold into 3-dimentional structures to achieve their physiological functions. However, aberrant protein misfolding and aggregation lead to various protein conformational diseases, such as Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and cardiac amyloidosis etc.
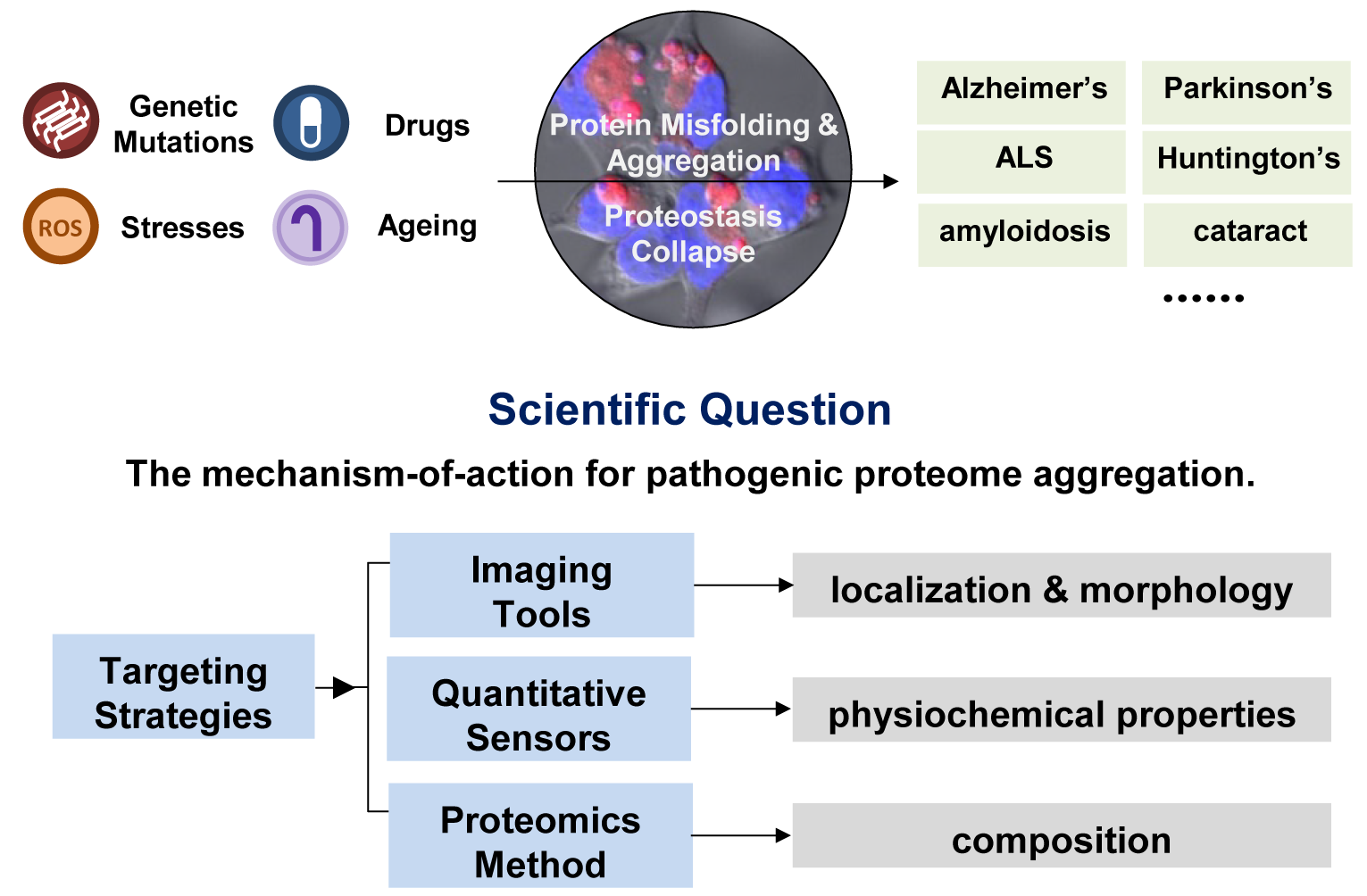
Direct visualization and quantitative analysis of aggregated proteins in living organisms are of great scientific significance for analyzing the occurrence and development mechanisms of related diseases. In particular, we are interested in revealing the localization, morphology, physicochemical properties, as well as compositions of aggregated proteins in stressed and diseased cells.
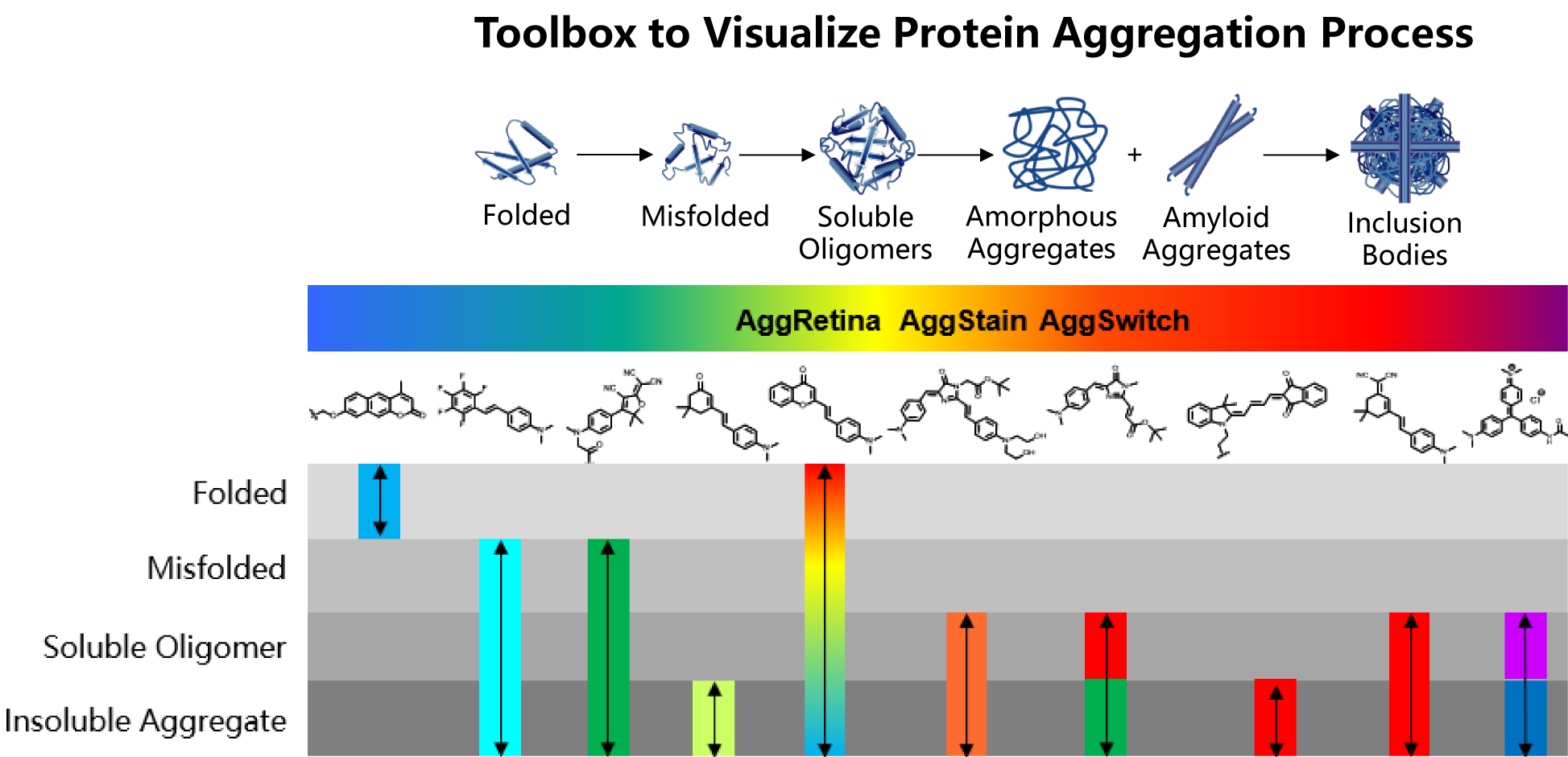
Towards these goals, our group focuses on developing novel chemical targeting strategies for aggregated proteome without defined 3-D structural guidance. Taking advantage of the entropy-driven hydrophobic effect of protein aggregation, we selectively target and distinguish protein misfolding and aggregation states by controlling targeting probes’ amphiphilicity, H-bonding acceptor/donor, aromaticity etc. For imaging tools, we regulated probes’ fluorescence environment-sensitivity to monitor the aggregation process. For proteomics analysis, we develop chemical proteomics probe to capture aggregated proteome in situ with spatiotemporal resolution.
Currently our lab works on these major projects:
1. Fluorescent probes to observe the protein aggregation process;
2. Chemical and biological sensors to quantify physicochemical properties of aggregated proteins;
3. Proteomics probes to analyze the components of the aggregated proteome;
4. Early diagnosis of transthyretin (ATTR) amyloid cardiomyopathy.